12 Tidy Data - R for Data Science
This post covers the content and exercises for Ch 12: Tidy Data from R for Data Science. The chapter teaches how to apply the organizational structure of tidy data to achieve a consistent format for data.
library(tidyverse)12.2 Tidy data
The concept of tidy data allows for a consistent format for data. The simple practical instruction for achieving tidy data are: 1. Put each dataset in a tibble. 2. Put each variable in a column.
12.2.1 Exercises
- Using prose, describe how the variables and observations are organised in each of the sample tables.
# table1
# > # A tibble: 6 x 4
# > country year cases population
# > <chr> <int> <int> <int>
# > 1 Afghanistan 1999 745 19987071
# > 2 Afghanistan 2000 2666 20595360
# > 3 Brazil 1999 37737 172006362
# > 4 Brazil 2000 80488 174504898
# > 5 China 1999 212258 1272915272
# > 6 China 2000 213766 1280428583table1is in tidy format. The 4 variables are the columns and each observation is a row.
# table2
#> # A tibble: 12 x 4
#> country year type count
#> <chr> <int> <chr> <int>
#> 1 Afghanistan 1999 cases 745
#> 2 Afghanistan 1999 population 19987071
#> 3 Afghanistan 2000 cases 2666
#> 4 Afghanistan 2000 population 20595360
#> 5 Brazil 1999 cases 37737
#> 6 Brazil 1999 population 172006362
#> # ... with 6 more rowstable2has two variables combined into a single variabletype. This means there are twice as many observations as with the tidy data.
# table3
#> # A tibble: 6 x 3
#> country year rate
#> * <chr> <int> <chr>
#> 1 Afghanistan 1999 745/19987071
#> 2 Afghanistan 2000 2666/20595360
#> 3 Brazil 1999 37737/172006362
#> 4 Brazil 2000 80488/174504898
#> 5 China 1999 212258/1272915272
#> 6 China 2000 213766/1280428583table3combines thecountvariable intorateand stores it as a character to maintain to two values forcasesandpopulationin the single variable. The labels for those values are no longer in the data.
# Spread across two tibbles
# table4a # cases
#> # A tibble: 3 x 3
#> country `1999` `2000`
#> * <chr> <int> <int>
#> 1 Afghanistan 745 2666
#> 2 Brazil 37737 80488
#> 3 China 212258 213766
# table4b # population
#> # A tibble: 3 x 3
#> country `1999` `2000`
#> * <chr> <int> <int>
#> 1 Afghanistan 19987071 20595360
#> 2 Brazil 172006362 174504898
#> 3 China 1272915272 1280428583table4splits the data into two different tables,aandb. In these tables the years are split into two different variables. There is an observation for each country in these tables.
Compute the rate for table2, and table4a + table4b. You will need to perform four operations:
- Extract the number of TB cases per country per year.
- Extract the matching population per country per year.
- Divide cases by population, and multiply by 10000.
- Store back in the appropriate place.
Which representation is easiest to work with? Which is hardest? Why?
table2 <- data_frame(country = c(rep("Afghanistan", 4), rep("Brazil", 4), rep("China", 4)),
year = rep(c(1999, 1999, 2000, 2000), 3),
type = rep(c("cases", "population"), 6),
count = c(745, 19987071, 2666, 20595360, 37737, 172006362, 80488, 174504898, 212258, 1272915272, 213766, 1280428583))## Warning: `data_frame()` is deprecated, use `tibble()`.
## This warning is displayed once per session.table2## # A tibble: 12 x 4
## country year type count
## <chr> <dbl> <chr> <dbl>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 1999 population 19987071
## 3 Afghanistan 2000 cases 2666
## 4 Afghanistan 2000 population 20595360
## 5 Brazil 1999 cases 37737
## 6 Brazil 1999 population 172006362
## 7 Brazil 2000 cases 80488
## 8 Brazil 2000 population 174504898
## 9 China 1999 cases 212258
## 10 China 1999 population 1272915272
## 11 China 2000 cases 213766
## 12 China 2000 population 1280428583(tb_cases <- table2 %>%
filter(type %in% "cases"))## # A tibble: 6 x 4
## country year type count
## <chr> <dbl> <chr> <dbl>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 2000 cases 2666
## 3 Brazil 1999 cases 37737
## 4 Brazil 2000 cases 80488
## 5 China 1999 cases 212258
## 6 China 2000 cases 213766(tb_pop <- table2 %>%
filter(type %in% "population"))## # A tibble: 6 x 4
## country year type count
## <chr> <dbl> <chr> <dbl>
## 1 Afghanistan 1999 population 19987071
## 2 Afghanistan 2000 population 20595360
## 3 Brazil 1999 population 172006362
## 4 Brazil 2000 population 174504898
## 5 China 1999 population 1272915272
## 6 China 2000 population 1280428583rate <- tb_cases
rate$rate <- tb_cases$count / tb_pop$count * 10000
rate## # A tibble: 6 x 5
## country year type count rate
## <chr> <dbl> <chr> <dbl> <dbl>
## 1 Afghanistan 1999 cases 745 0.373
## 2 Afghanistan 2000 cases 2666 1.29
## 3 Brazil 1999 cases 37737 2.19
## 4 Brazil 2000 cases 80488 4.61
## 5 China 1999 cases 212258 1.67
## 6 China 2000 cases 213766 1.67- Recreate the plot showing change in cases over time using table2 instead of table1. What do you need to do first?
table2 %>%
filter(type %in% "cases") %>%
ggplot(aes(x = year, y = count)) +
geom_point(aes(color = country), size = 3) +
geom_line(aes(group = country))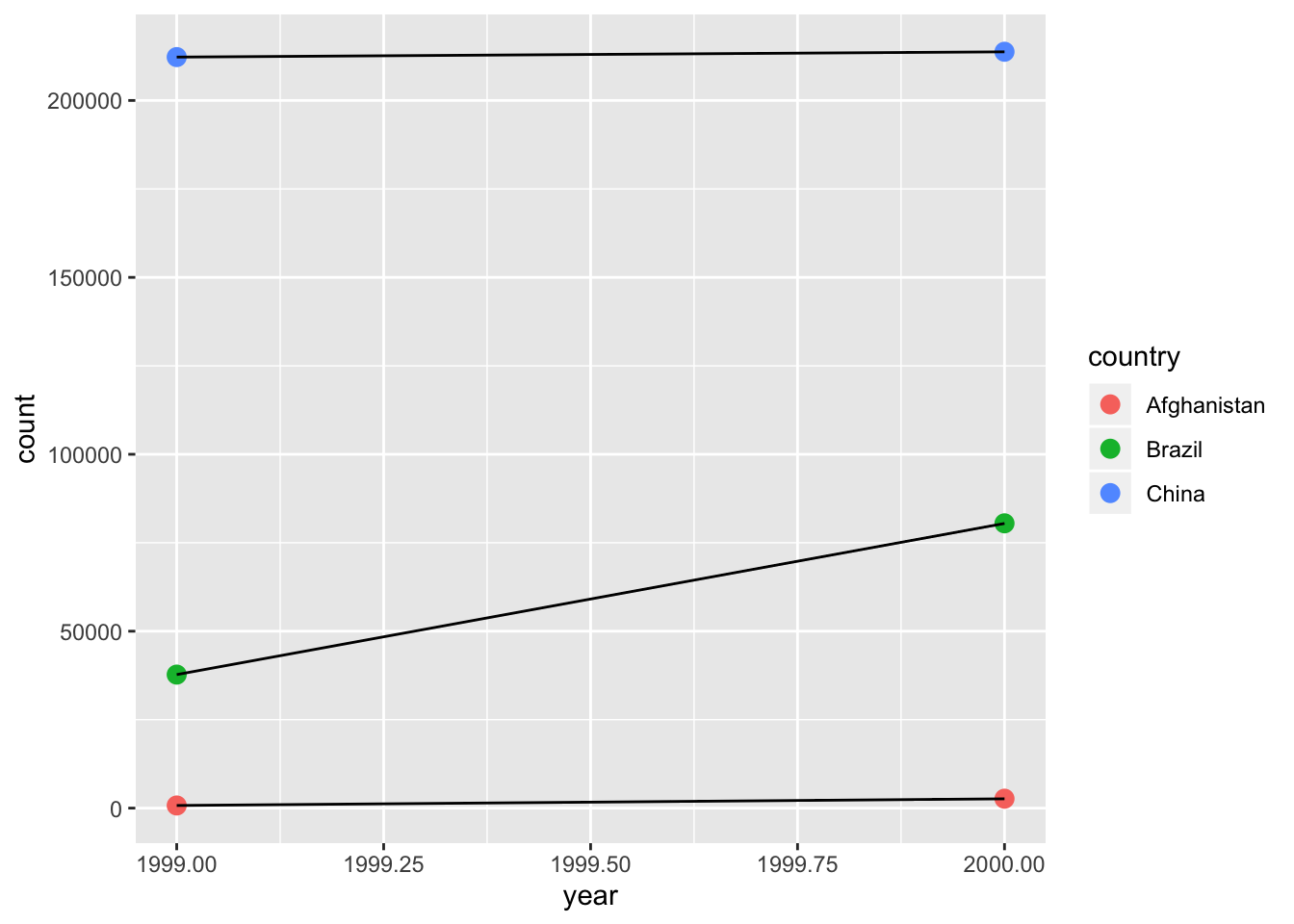
- First you need to filter
typeforcases.
12.3 Spreading and gathering
Often need to resolve one of two common problems when tidying data:
One variable might be spread across multiple columns.
One observation might be scattered across multiple rows.
12.3.3 Exercises
- Why are gather() and spread() not perfectly symmetrical? Carefully consider the following example:
(stocks <- tibble(
year = c(2015, 2015, 2016, 2016),
half = c( 1, 2, 1, 2),
return = c(1.88, 0.59, 0.92, 0.17)
))## # A tibble: 4 x 3
## year half return
## <dbl> <dbl> <dbl>
## 1 2015 1 1.88
## 2 2015 2 0.59
## 3 2016 1 0.92
## 4 2016 2 0.17stocks %>%
spread(year, return) %>%
gather("year", "return", `2015`:`2016`)## # A tibble: 4 x 3
## half year return
## <dbl> <chr> <dbl>
## 1 1 2015 1.88
## 2 2 2015 0.59
## 3 1 2016 0.92
## 4 2 2016 0.17(Hint: look at the variable types and think about column names.)
Both spread() and gather() have a convert argument. What does it do?
- In the example
yearis converted tocharacterwhen it is gathered back after being spread. This happens becauseconvertis by defaultFALSEso it does not attempt to determine the appropriate class for new columns. Whenconvertis set toTRUE, the year is correctly converted to a number when gathered as seen in the example below.
stocks %>%
spread(year, return) %>%
gather("year", "return", `2015`:`2016`, convert = TRUE)## # A tibble: 4 x 3
## half year return
## <dbl> <int> <dbl>
## 1 1 2015 1.88
## 2 2 2015 0.59
## 3 1 2016 0.92
## 4 2 2016 0.17- Why does this code fail?
(table4a <- table2 %>%
filter(type %in% "cases") %>%
spread(year, count) %>%
select(-type))## # A tibble: 3 x 3
## country `1999` `2000`
## <chr> <dbl> <dbl>
## 1 Afghanistan 745 2666
## 2 Brazil 37737 80488
## 3 China 212258 213766table4a %>%
gather(1999, 2000, key = "year", value = "cases")## Error: Can't subset columns that don't exist.
## [31mx[39m The locations 1999 and 2000 don't exist.
## [34mℹ[39m There are only 3 columns.- Since the years are numbers they need to be selected with backticks as in the example below.
table4a %>%
gather(`1999`, `2000`, key = "year", value = "cases", convert = TRUE)## # A tibble: 6 x 3
## country year cases
## <chr> <int> <dbl>
## 1 Afghanistan 1999 745
## 2 Brazil 1999 37737
## 3 China 1999 212258
## 4 Afghanistan 2000 2666
## 5 Brazil 2000 80488
## 6 China 2000 213766- Why does spreading this tibble fail? How could you add a new column to fix the problem?
(people <- tribble(
~name, ~key, ~value,
#-----------------|--------|------
"Phillip Woods", "age", 45,
"Phillip Woods", "height", 186,
"Phillip Woods", "age", 50,
"Jessica Cordero", "age", 37,
"Jessica Cordero", "height", 156
))## # A tibble: 5 x 3
## name key value
## <chr> <chr> <dbl>
## 1 Phillip Woods age 45
## 2 Phillip Woods height 186
## 3 Phillip Woods age 50
## 4 Jessica Cordero age 37
## 5 Jessica Cordero height 156people %>%
spread(key, value)## Error: Each row of output must be identified by a unique combination of keys.
## Keys are shared for 2 rows:
## * 1, 3- It fails since there are two values for Phillip in age. Fix by adding an index.
(people <- tribble(
~name, ~key, ~value, ~index,
#-----------------|--------|--------|------
"Phillip Woods", "age", 45, 1,
"Phillip Woods", "height", 186, 1,
"Phillip Woods", "age", 50, 2,
"Jessica Cordero", "age", 37, 1,
"Jessica Cordero", "height", 156, 1
))## # A tibble: 5 x 4
## name key value index
## <chr> <chr> <dbl> <dbl>
## 1 Phillip Woods age 45 1
## 2 Phillip Woods height 186 1
## 3 Phillip Woods age 50 2
## 4 Jessica Cordero age 37 1
## 5 Jessica Cordero height 156 1people %>%
spread(key, value)## # A tibble: 3 x 4
## name index age height
## <chr> <dbl> <dbl> <dbl>
## 1 Jessica Cordero 1 37 156
## 2 Phillip Woods 1 45 186
## 3 Phillip Woods 2 50 NA- Tidy the simple tibble below. Do you need to spread or gather it? What are the variables?
preg <- tribble(
~pregnant, ~male, ~female,
"yes", NA, 10,
"no", 20, 12
)
preg %>%
gather(male, female, key = "sex", value = "count")## # A tibble: 4 x 3
## pregnant sex count
## <chr> <chr> <dbl>
## 1 yes male NA
## 2 no male 20
## 3 yes female 10
## 4 no female 1212.4 Separating and uniting
separate() is used when multiple variables are stored together in a single variable.
unite() is the inverse.
12.4.3 Exercises
- What do the extra and fill arguments do in separate()? Experiment with the various options for the following two toy datasets.
tibble(x = c("a,b,c", "d,e,f,g", "h,i,j")) %>%
separate(x, c("one", "two", "three"))## Warning: Expected 3 pieces. Additional pieces discarded in 1 rows [2].## # A tibble: 3 x 3
## one two three
## <chr> <chr> <chr>
## 1 a b c
## 2 d e f
## 3 h i jtibble(x = c("a,b,c", "d,e,f,g", "h,i,j")) %>%
separate(x, c("one", "two", "three"), extra = "drop")## # A tibble: 3 x 3
## one two three
## <chr> <chr> <chr>
## 1 a b c
## 2 d e f
## 3 h i jtibble(x = c("a,b,c", "d,e,f,g", "h,i,j")) %>%
separate(x, c("one", "two", "three"), extra = "merge")## # A tibble: 3 x 3
## one two three
## <chr> <chr> <chr>
## 1 a b c
## 2 d e f,g
## 3 h i jtibble(x = c("a,b,c", "d,e", "f,g,i")) %>%
separate(x, c("one", "two", "three"))## Warning: Expected 3 pieces. Missing pieces filled with `NA` in 1 rows [2].## # A tibble: 3 x 3
## one two three
## <chr> <chr> <chr>
## 1 a b c
## 2 d e <NA>
## 3 f g itibble(x = c("a,b,c", "d,e", "f,g,i")) %>%
separate(x, c("one", "two", "three"), fill = "right")## # A tibble: 3 x 3
## one two three
## <chr> <chr> <chr>
## 1 a b c
## 2 d e <NA>
## 3 f g itibble(x = c("a,b,c", "d,e", "f,g,i")) %>%
separate(x, c("one", "two", "three"), fill = "left")## # A tibble: 3 x 3
## one two three
## <chr> <chr> <chr>
## 1 a b c
## 2 <NA> d e
## 3 f g i- They control the behavior for when a vector has too many or too few pieces.
- Both unite() and separate() have a remove argument. What does it do? Why would you set it to FALSE?
- It removes the original column that was modified. Setting to
FALSEwould allow you to keep the original variable in the data.
- Compare and contrast separate() and extract(). Why are there three variations of separation (by position, by separator, and with groups), but only one unite?
extract()separates on a defined regex. There is only one variation forunite()since the columns are already specified and it only requires a separator to be defined. When separating it is more ambiguous so more options are required to cover possibilities.
12.5 Missing values
Can be:
* Explicit: flagged with NA
* Implicit: not present in data
12.5.1 Exercises
- Compare and contrast the fill arguments to spread() and complete().
spread()will replaceNAwith whatever is specified infillcomplete()A named list that for each variable supplies a single value to use instead of NA for missing combinations. Like withspread()if specified will replace both explicit and implicitNA
- What does the direction argument to fill() do?
- Changes which direction to use for the previous entry, default to down.
12.6 Case Study
who## # A tibble: 7,240 x 60
## country iso2 iso3 year new_sp_m014 new_sp_m1524 new_sp_m2534 new_sp_m3544
## <chr> <chr> <chr> <int> <int> <int> <int> <int>
## 1 Afghan… AF AFG 1980 NA NA NA NA
## 2 Afghan… AF AFG 1981 NA NA NA NA
## 3 Afghan… AF AFG 1982 NA NA NA NA
## 4 Afghan… AF AFG 1983 NA NA NA NA
## 5 Afghan… AF AFG 1984 NA NA NA NA
## 6 Afghan… AF AFG 1985 NA NA NA NA
## 7 Afghan… AF AFG 1986 NA NA NA NA
## 8 Afghan… AF AFG 1987 NA NA NA NA
## 9 Afghan… AF AFG 1988 NA NA NA NA
## 10 Afghan… AF AFG 1989 NA NA NA NA
## # … with 7,230 more rows, and 52 more variables: new_sp_m4554 <int>,
## # new_sp_m5564 <int>, new_sp_m65 <int>, new_sp_f014 <int>,
## # new_sp_f1524 <int>, new_sp_f2534 <int>, new_sp_f3544 <int>,
## # new_sp_f4554 <int>, new_sp_f5564 <int>, new_sp_f65 <int>,
## # new_sn_m014 <int>, new_sn_m1524 <int>, new_sn_m2534 <int>,
## # new_sn_m3544 <int>, new_sn_m4554 <int>, new_sn_m5564 <int>,
## # new_sn_m65 <int>, new_sn_f014 <int>, new_sn_f1524 <int>,
## # new_sn_f2534 <int>, new_sn_f3544 <int>, new_sn_f4554 <int>,
## # new_sn_f5564 <int>, new_sn_f65 <int>, new_ep_m014 <int>,
## # new_ep_m1524 <int>, new_ep_m2534 <int>, new_ep_m3544 <int>,
## # new_ep_m4554 <int>, new_ep_m5564 <int>, new_ep_m65 <int>,
## # new_ep_f014 <int>, new_ep_f1524 <int>, new_ep_f2534 <int>,
## # new_ep_f3544 <int>, new_ep_f4554 <int>, new_ep_f5564 <int>,
## # new_ep_f65 <int>, newrel_m014 <int>, newrel_m1524 <int>,
## # newrel_m2534 <int>, newrel_m3544 <int>, newrel_m4554 <int>,
## # newrel_m5564 <int>, newrel_m65 <int>, newrel_f014 <int>,
## # newrel_f1524 <int>, newrel_f2534 <int>, newrel_f3544 <int>,
## # newrel_f4554 <int>, newrel_f5564 <int>, newrel_f65 <int>(who1 <- who %>%
gather(new_sp_m014:newrel_f65, key = "key", value = "cases", na.rm = TRUE))## # A tibble: 76,046 x 6
## country iso2 iso3 year key cases
## <chr> <chr> <chr> <int> <chr> <int>
## 1 Afghanistan AF AFG 1997 new_sp_m014 0
## 2 Afghanistan AF AFG 1998 new_sp_m014 30
## 3 Afghanistan AF AFG 1999 new_sp_m014 8
## 4 Afghanistan AF AFG 2000 new_sp_m014 52
## 5 Afghanistan AF AFG 2001 new_sp_m014 129
## 6 Afghanistan AF AFG 2002 new_sp_m014 90
## 7 Afghanistan AF AFG 2003 new_sp_m014 127
## 8 Afghanistan AF AFG 2004 new_sp_m014 139
## 9 Afghanistan AF AFG 2005 new_sp_m014 151
## 10 Afghanistan AF AFG 2006 new_sp_m014 193
## # … with 76,036 more rows(who2 <- who1 %>%
mutate(key = stringr::str_replace(key, "newrel", "new_rel")))## # A tibble: 76,046 x 6
## country iso2 iso3 year key cases
## <chr> <chr> <chr> <int> <chr> <int>
## 1 Afghanistan AF AFG 1997 new_sp_m014 0
## 2 Afghanistan AF AFG 1998 new_sp_m014 30
## 3 Afghanistan AF AFG 1999 new_sp_m014 8
## 4 Afghanistan AF AFG 2000 new_sp_m014 52
## 5 Afghanistan AF AFG 2001 new_sp_m014 129
## 6 Afghanistan AF AFG 2002 new_sp_m014 90
## 7 Afghanistan AF AFG 2003 new_sp_m014 127
## 8 Afghanistan AF AFG 2004 new_sp_m014 139
## 9 Afghanistan AF AFG 2005 new_sp_m014 151
## 10 Afghanistan AF AFG 2006 new_sp_m014 193
## # … with 76,036 more rows(who3 <- who2 %>%
separate(key, c("new", "type", "sexage"), sep = "_"))## # A tibble: 76,046 x 8
## country iso2 iso3 year new type sexage cases
## <chr> <chr> <chr> <int> <chr> <chr> <chr> <int>
## 1 Afghanistan AF AFG 1997 new sp m014 0
## 2 Afghanistan AF AFG 1998 new sp m014 30
## 3 Afghanistan AF AFG 1999 new sp m014 8
## 4 Afghanistan AF AFG 2000 new sp m014 52
## 5 Afghanistan AF AFG 2001 new sp m014 129
## 6 Afghanistan AF AFG 2002 new sp m014 90
## 7 Afghanistan AF AFG 2003 new sp m014 127
## 8 Afghanistan AF AFG 2004 new sp m014 139
## 9 Afghanistan AF AFG 2005 new sp m014 151
## 10 Afghanistan AF AFG 2006 new sp m014 193
## # … with 76,036 more rows(who4 <- who3 %>%
select(-new, -iso2, -iso3))## # A tibble: 76,046 x 5
## country year type sexage cases
## <chr> <int> <chr> <chr> <int>
## 1 Afghanistan 1997 sp m014 0
## 2 Afghanistan 1998 sp m014 30
## 3 Afghanistan 1999 sp m014 8
## 4 Afghanistan 2000 sp m014 52
## 5 Afghanistan 2001 sp m014 129
## 6 Afghanistan 2002 sp m014 90
## 7 Afghanistan 2003 sp m014 127
## 8 Afghanistan 2004 sp m014 139
## 9 Afghanistan 2005 sp m014 151
## 10 Afghanistan 2006 sp m014 193
## # … with 76,036 more rows(who5 <- who4 %>%
separate(sexage, c("sex", "age"), sep = 1))## # A tibble: 76,046 x 6
## country year type sex age cases
## <chr> <int> <chr> <chr> <chr> <int>
## 1 Afghanistan 1997 sp m 014 0
## 2 Afghanistan 1998 sp m 014 30
## 3 Afghanistan 1999 sp m 014 8
## 4 Afghanistan 2000 sp m 014 52
## 5 Afghanistan 2001 sp m 014 129
## 6 Afghanistan 2002 sp m 014 90
## 7 Afghanistan 2003 sp m 014 127
## 8 Afghanistan 2004 sp m 014 139
## 9 Afghanistan 2005 sp m 014 151
## 10 Afghanistan 2006 sp m 014 193
## # … with 76,036 more rows12.6.1 Exercises
- In this case study I set na.rm = TRUE just to make it easier to check that we had the correct values. Is this reasonable? Think about how missing values are represented in this dataset. Are there implicit missing values? What’s the difference between an NA and zero?
- In this data
NAis assigned to values when there wasn’t a recording for that case in a particular year. A 0 is assigned if the data was recorded but there were no cases. Since there are no implicit missing values it is okay to dropNA.
who %>%
count(country) %>%
arrange(n) ## # A tibble: 219 x 2
## country n
## <chr> <int>
## 1 South Sudan 3
## 2 Bonaire, Saint Eustatius and Saba 4
## 3 Curacao 4
## 4 Sint Maarten (Dutch part) 4
## 5 Montenegro 9
## 6 Serbia 9
## 7 Timor-Leste 12
## 8 Serbia & Montenegro 25
## 9 Netherlands Antilles 30
## 10 Afghanistan 34
## # … with 209 more rowswho %>%
filter(country %in% c("South Sudan",
"Bonaire, Saint Eustatius and Saba"))## # A tibble: 7 x 60
## country iso2 iso3 year new_sp_m014 new_sp_m1524 new_sp_m2534 new_sp_m3544
## <chr> <chr> <chr> <int> <int> <int> <int> <int>
## 1 Bonair… BQ BES 2010 0 0 0 0
## 2 Bonair… BQ BES 2011 0 0 0 0
## 3 Bonair… BQ BES 2012 0 0 0 0
## 4 Bonair… BQ BES 2013 NA NA NA NA
## 5 South … SS SSD 2011 39 251 599 402
## 6 South … SS SSD 2012 42 356 753 462
## 7 South … SS SSD 2013 NA NA NA NA
## # … with 52 more variables: new_sp_m4554 <int>, new_sp_m5564 <int>,
## # new_sp_m65 <int>, new_sp_f014 <int>, new_sp_f1524 <int>,
## # new_sp_f2534 <int>, new_sp_f3544 <int>, new_sp_f4554 <int>,
## # new_sp_f5564 <int>, new_sp_f65 <int>, new_sn_m014 <int>,
## # new_sn_m1524 <int>, new_sn_m2534 <int>, new_sn_m3544 <int>,
## # new_sn_m4554 <int>, new_sn_m5564 <int>, new_sn_m65 <int>,
## # new_sn_f014 <int>, new_sn_f1524 <int>, new_sn_f2534 <int>,
## # new_sn_f3544 <int>, new_sn_f4554 <int>, new_sn_f5564 <int>,
## # new_sn_f65 <int>, new_ep_m014 <int>, new_ep_m1524 <int>,
## # new_ep_m2534 <int>, new_ep_m3544 <int>, new_ep_m4554 <int>,
## # new_ep_m5564 <int>, new_ep_m65 <int>, new_ep_f014 <int>,
## # new_ep_f1524 <int>, new_ep_f2534 <int>, new_ep_f3544 <int>,
## # new_ep_f4554 <int>, new_ep_f5564 <int>, new_ep_f65 <int>,
## # newrel_m014 <int>, newrel_m1524 <int>, newrel_m2534 <int>,
## # newrel_m3544 <int>, newrel_m4554 <int>, newrel_m5564 <int>,
## # newrel_m65 <int>, newrel_f014 <int>, newrel_f1524 <int>,
## # newrel_f2534 <int>, newrel_f3544 <int>, newrel_f4554 <int>,
## # newrel_f5564 <int>, newrel_f65 <int>- What happens if you neglect the mutate() step? (mutate(key = stringr::str_replace(key, “newrel”, “new_rel”)))
- It doesn’t know to separate the values stored as
newreland ends up giving a too few values warning.
who %>%
gather(code, value, new_sp_m014:newrel_f65, na.rm = TRUE) %>%
separate(code, c("new", "var", "sexage")) %>%
select(-new, -iso2, -iso3) %>%
separate(sexage, c("sex", "age"), sep = 1)## Warning: Expected 3 pieces. Missing pieces filled with `NA` in 2580 rows [73467,
## 73468, 73469, 73470, 73471, 73472, 73473, 73474, 73475, 73476, 73477, 73478,
## 73479, 73480, 73481, 73482, 73483, 73484, 73485, 73486, ...].## # A tibble: 76,046 x 6
## country year var sex age value
## <chr> <int> <chr> <chr> <chr> <int>
## 1 Afghanistan 1997 sp m 014 0
## 2 Afghanistan 1998 sp m 014 30
## 3 Afghanistan 1999 sp m 014 8
## 4 Afghanistan 2000 sp m 014 52
## 5 Afghanistan 2001 sp m 014 129
## 6 Afghanistan 2002 sp m 014 90
## 7 Afghanistan 2003 sp m 014 127
## 8 Afghanistan 2004 sp m 014 139
## 9 Afghanistan 2005 sp m 014 151
## 10 Afghanistan 2006 sp m 014 193
## # … with 76,036 more rows- I claimed that iso2 and iso3 were redundant with country. Confirm this claim.
- There are no additional rows when adding the iso variables to the grouping.
who %>%
count(country)## # A tibble: 219 x 2
## country n
## <chr> <int>
## 1 Afghanistan 34
## 2 Albania 34
## 3 Algeria 34
## 4 American Samoa 34
## 5 Andorra 34
## 6 Angola 34
## 7 Anguilla 34
## 8 Antigua and Barbuda 34
## 9 Argentina 34
## 10 Armenia 34
## # … with 209 more rowswho %>%
count(country, iso2, iso3)## # A tibble: 219 x 4
## country iso2 iso3 n
## <chr> <chr> <chr> <int>
## 1 Afghanistan AF AFG 34
## 2 Albania AL ALB 34
## 3 Algeria DZ DZA 34
## 4 American Samoa AS ASM 34
## 5 Andorra AD AND 34
## 6 Angola AO AGO 34
## 7 Anguilla AI AIA 34
## 8 Antigua and Barbuda AG ATG 34
## 9 Argentina AR ARG 34
## 10 Armenia AM ARM 34
## # … with 209 more rows- For each country, year, and sex compute the total number of cases of TB. Make an informative visualisation of the data.
who5 %>%
filter(sex %in% "f") %>%
ggplot(aes(x = year, y = country)) +
geom_tile(aes(fill = cases)) 
Not actually an informative visualization yet.